GitHub - schatzlab/crossstitch: Code for phasing SVs with SNPs
Code for phasing SVs with SNPs. Contribute to schatzlab/crossstitch development by creating an account on GitHub.
GitHub - PhysiologicAILab/SAM-CL: Self-adversarial Multi-scale Contrastive Learning for Semantic Segmentation of Thermal Facial Images
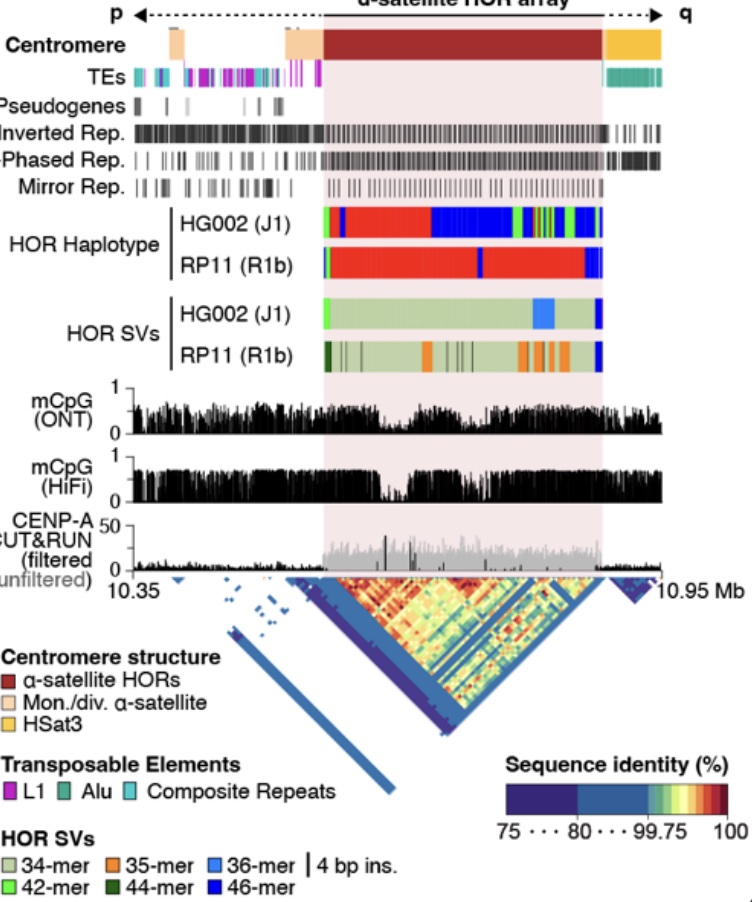
Schatzlab - Publications

How to add comparissons between treatments for heach gene expression in the same plot? ggboxplot · Issue #169 · kassambara/ggpubr · GitHub

Semi-automated assembly of high-quality diploid human reference genomes. - Abstract - Europe PMC
Heterozygous SVs and homozygous SVs · Issue #72 · fritzsedlazeck/Sniffles · GitHub
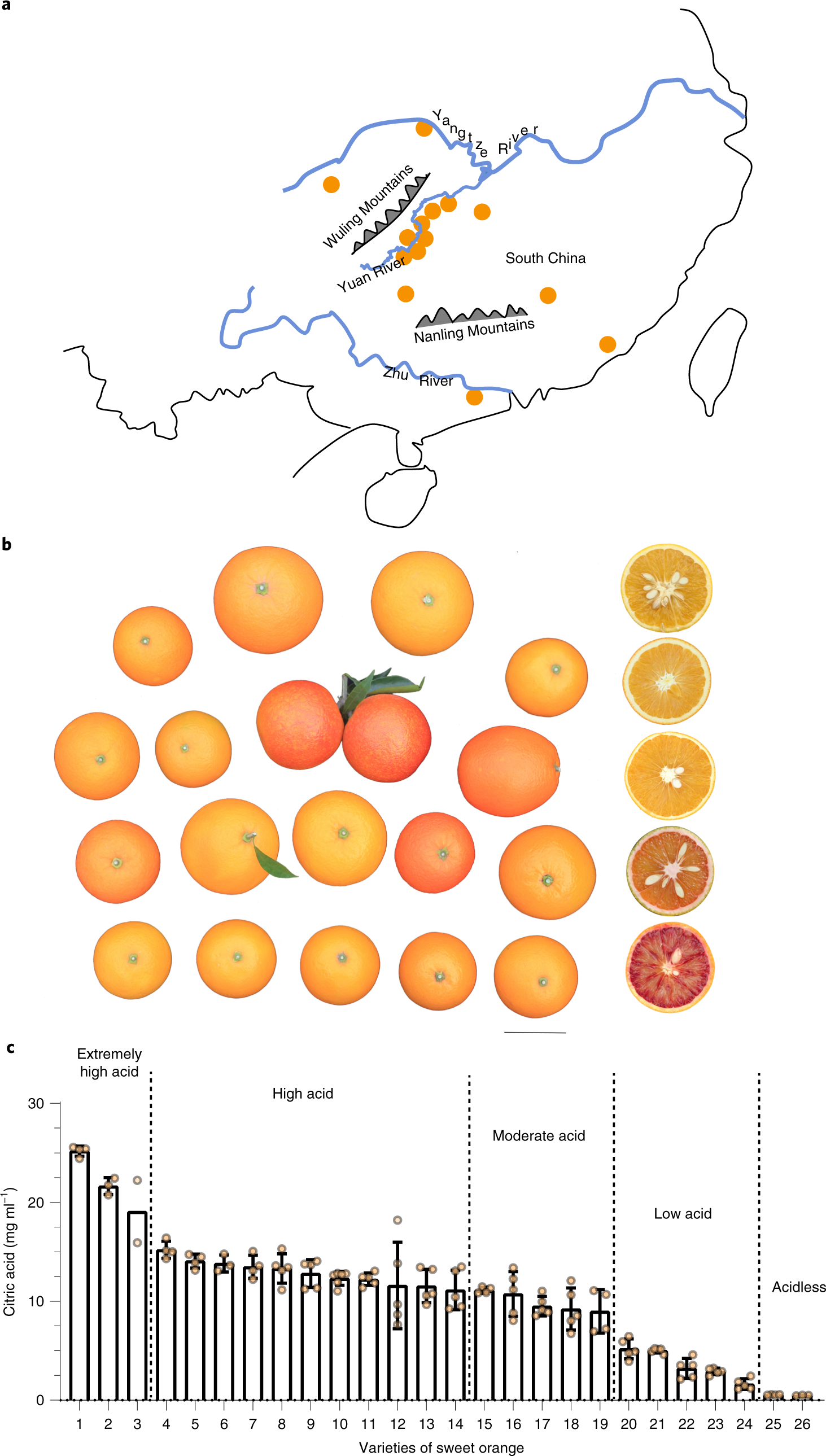
Somatic variations led to the selection of acidic and acidless orange cultivars

Paragraph: A graph-based structural variant genotyper for short-read sequence data
Squiz.NamingConventions.ValidVariableName how to only report for declaration, not usage · Issue #3714 · squizlabs/PHP_CodeSniffer · GitHub

Automated assembly of high-quality diploid human reference genomes

Publications — Salzman Lab
GitHub - apriha/snps: tools for reading, writing, merging, and remapping SNPs
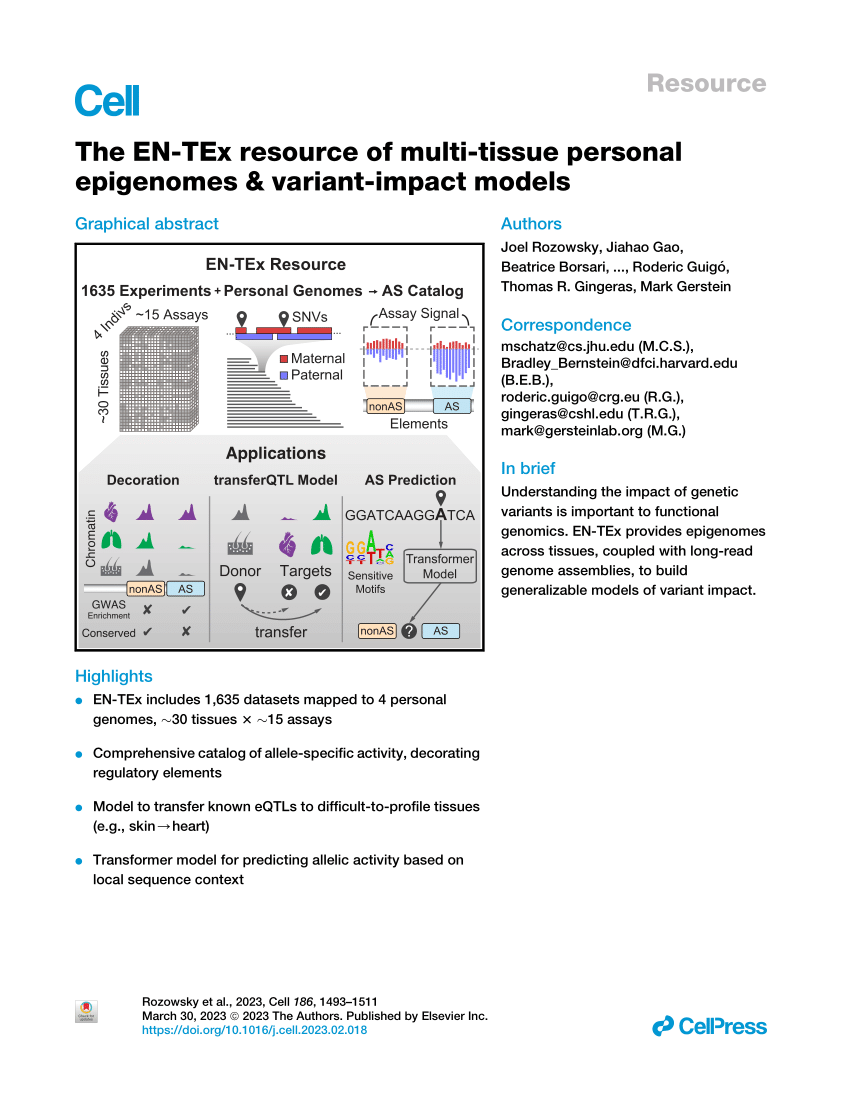
PDF) The EN-TEx resource of multi-tissue personal epigenomes & variant-impact models